作者:Shen,ZJ; Chen, J; Ghoto, K; Hu, WJ; Gao, GF; Luo, MR; Li, Z; Simon, M; Zhu, XY;Zheng, HL
影响因子:3.39
刊物名称:TREE PHYSIOLOGY
出版年份:2018
卷:38 页码:1605-622
Avicennia marina(Forsk.) Vierh is one of the most salt-tolerant mangrove species. Our previous study revealed that nitric oxide (NO)enhanced the salt tolerance ofA. marinaby promoting salt secretion and Na+sequestration under salt stress. However, little isknown about the regulation of NO on proteomic profiling for this mangrove species. In this study, we used sodium nitroprusside(SNP), an NO donor, to investigate the regulatory mechanism of NO on salt tolerance ofA. marinaaccording to physiological andproteomic aspects.
Photosynthesis data showed that the reduction in photosynthesis caused by high salinity treatment (400 mMNaCl) could be partially recovered by addition of SNP (100μM). Further analysis revealed that the high salinity treatment couldinduce not only the stomatal limitation but also non-stomatal limitation on photosynthetic reduction, while SNP addition couldrestore the non-stomatal limitation, implying that the application of SNP was beneficial to the metabolic process in leaves. Proteomicanalysis identified 49 differentially expressed
proteins involved in various biological processes such as photosynthesis, energy metabolism, primary metabolism, RNA transcription, protein translation and stress response proteins. Under high salinity treatment, theabundances of proteins related to photosynthesis, such as ribulose-phosphate 3-epimerase (RPE, spot 3), RuBisCO large subunit(RBCL, spot 4, 5, 24), RuBisCO activase A (RCA, spot 17, 18) and quinine oxidoreductase-like protein isoform 1 (QOR1, spot 23),were significantly decreased. However, the abundance of proteins such as RBCL (spot 5, 9) and QOR1 (spot 23) were increased bySNP addition. In addition, exogenous NO supply alleviated salt tolerance by increasing the accumulation of some proteins involved in energy metabolism (spot 15), primary metabolism (spot 25, 45, 46), RNA transcription (spot 36) and stress response proteins (spot12, 21, 26, 37, 43). The
transcriptional levels of nine selected proteins were mostly consistent with their protein abundance exceptspot 46. Overall, the presented data demonstrated that NO has a positive effect on improving salt tolerance inA. marinaby regulating the protein abundance involved in photosynthesis, energy metabolism, primary metabolism and stress response.
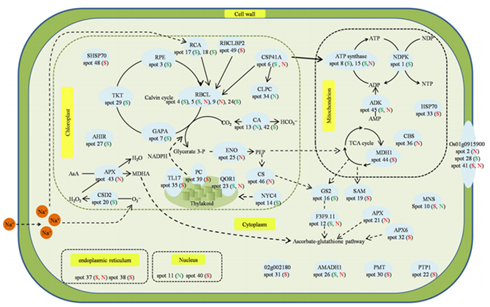
Figure 8. The proposed regulatory networks of NO on salt tolerance ofA. marinaleaves. The‘S’or‘N’in brackets stand for high salinity or SNP addition,respectively. The red color
of‘S’or‘N’indicates the up-regulated change, the green color of‘S’or‘N’indicates the down-regulated change. ENO: enolase, TKT: transketolase, RPE: ribulose-phosphate 3-epimerase, MDH1: malate dehydrogenase, HSP: heat shock protein, CSP41: chloroplast stem-loopbinding protein-41, GAPA:glyceraldehyde-3-phosphate ehydrogenase A, MDH1: malate dehydrogenase, RBCL: RuBisCO large subunit, AHIR: acetohydroxyacid isomeroreductase, MNS: lysosomal alpha-mannosidase, TL17: thylakoid lumenal 17.4 kDa protein, CA: carbonic anhydrase, NYC4:Os07g0558500, PC: plastocyanin, QOR1: quinone oxidoreductase-like protein isoform 1,AMADH1: aminoaldehyde dehydrogenase 1, PMT: probablemethyltransferase PMT11-like, 02g002180: hypothetical protein SORBIDRAFT_02g002180, APX: L-ascorbate peroxidase, APX6: L-ascorbate peroxidase T isoform 6, F3F9.11: F3F9.11, SHSP70: stromal 70 kDa heat shock-related protein, RBCLBP2: RuBisCO large subunit-binding protein subunitalphaisoform 2, PTP1: phosphate transporter traffic facilitator1 isoform 1, RCA: RuBisCO activase A. The detailed information for each spot is shown inTable1.

